Capsid Integrity qPCR—An Azo-Dye Based and Culture-Independent Approach to Estimate Adenovirus Infectivity after Disinfection and in the Aquatic Environment
Abstract
:1. Introduction
2. Material and Methods
2.1. Propagation and Enumeration of Virus Stocks
2.2. Thermal, UV, and Hypochlorite Inactivation
2.3. Collection and Concentration of Water Samples for Virus Analysis
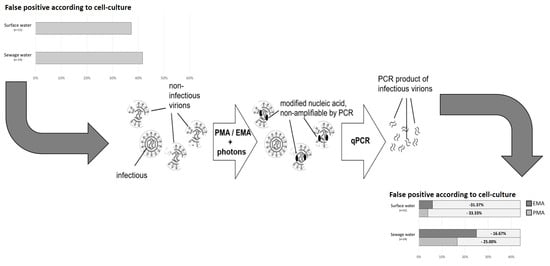
2.4. Dye Pretreatment
2.5. Extraction of Viral DNA and Quantification of Adenovirus Genomes
3. Statistical Analysis
4. Results and Discussion
Correlation between qPCR, ci-qPCR and Cell Culture in Environmental and Spike Samples
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stazi, V.; Tomei, M.C. Enhancing anaerobic treatment of domestic wastewater: State of the art, innovative technologies and future perspectives. Sci. Total Environ. 2018, 635, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Enger, K.S.; Nelson, K.L.; Clasen, T.; Rose, J.B.; Eisenberg, J.N. Linking quantitative microbial risk assessment and epidemiological data: Informing safe drinking water trials in developing countries. Environ. Sci. Technol. 2012, 46, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
- Chahal, C.; van den Akker, B.; Young, F.; Franco, C.; Blackbeard, J.; Monis, P. Pathogen and Particle Associations in Wastewater: Significance and Implications for Treatment and Disinfection Processes. Adv. Appl. Microbiol. 2016, 97, 63–119. [Google Scholar] [PubMed]
- Ngwenya, N.; Ncube, E.J.; Parsons, J. Recent advances in drinking water disinfection: Successes and challenges. Rev. Environ. Contam Toxicol. 2013, 222, 111–170. [Google Scholar] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2012; Volume 22, p. 1496. [Google Scholar]
- WHO. Guidelines for Safe Recreational Water Environments. Vol. 1: Coastal and Fresh Waters; World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Rames, E.; Roiko, A.; Stratton, H.; Macdonald, J. Technical aspects of using human adenovirus as a viral water quality indicator. Water Res. 2016, 96, 308–326. [Google Scholar] [CrossRef]
- Dias, E.; Ebdon, J.; Taylor, H. The application of bacteriophages as novel indicators of viral pathogens in wastewater treatment systems. Water Res. 2018, 129, 172–179. [Google Scholar] [CrossRef]
- McMinn, B.R.; Korajkic, A.; Ashbolt, N.J. Evaluation of Bacteroides fragilis GB-124 bacteriophages as novel human-associated faecal indicators in the United States. Lett. Appl. Microbiol. 2014, 59, 115–121. [Google Scholar] [CrossRef]
- Beck, S.E.; Hull, N.M.; Poepping, C.; Linden, K.G. Wavelength-dependent damage to adenoviral proteins across the germicidal UV spectrum. Environ. Sci. Technol. 2018, 52, 223–229. [Google Scholar] [CrossRef]
- Sinclair, R.G.; Jones, E.L.; Gerba, C.P. Viruses in recreational water-borne disease outbreaks: A review. J. Appl. Microbiol. 2009, 107, 1769–1780. [Google Scholar] [CrossRef]
- Soller, J.A.; Schoen, M.E.; Bartrand, T.; Ravenscroft, J.E.; Ashbolt, N.J. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 2010, 44, 4674–4691. [Google Scholar] [CrossRef]
- Ashbolt, N.J. Microbial Contamination of Drinking Water and Human Health from Community Water Systems. Curr. Environ. Health Rep. 2015, 2, 95–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassebaum, N.J.; Bertozzi-Villa, A.; Coggeshall, M.S.; Shackelford, K.A.; Steiner, C.; Heuton, K.R.; Gonzalez-Medina, D.; Barber, R.; Huynh, C.; Dicker, D.; et al. Global, regional, and national levels and causes of maternal mortality during 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 980–1004. [Google Scholar] [CrossRef]
- Gerba, C.P.; Betancourt, W.Q.; Kitajima, M.; Rock, C.M. Reducing uncertainty in estimating virus reduction by advanced water treatment processes. Water Res. 2018, 133, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Beck, S.E.; Rodriguez, R.A.; Linden, K.G.; Hargy, T.M.; Larason, T.C.; Wright, H.B. Wavelength dependent UV inactivation and DNA damage of adenovirus as measured by cell culture infectivity and long range quantitative PCR. Environ. Sci. Technol. 2014, 48, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Gall, A.M.; Marinas, B.J.; Lu, Y.; Shisler, J.L. Waterborne Viruses: A Barrier to Safe Drinking Water. PLoS Pathog. 2015, 11, e1004867. [Google Scholar] [CrossRef]
- Gerba, C.P.; Betancourt, W.Q. Viral Aggregation: Impact on Virus Behavior in the Environment. Environ. Sci. Technol. 2017, 51, 7318–7325. [Google Scholar] [CrossRef]
- Clasen, T.; Pruss-Ustun, A.; Mathers, C.D.; Cumming, O.; Cairncross, S.; Colford, J.M., Jr. Estimating the impact of unsafe water, sanitation and hygiene on the global burden of disease: Evolving and alternative methods. Trop. Med. Int. Health 2014, 19, 884–893. [Google Scholar] [CrossRef]
- Farkas, K.; Marshall, M.; Cooper, D.; McDonald, J.E.; Malham, S.K.; Peters, D.E.; Maloney, J.D.; Jones, D.L. Seasonal and diurnal surveillance of treated and untreated wastewater for human enteric viruses. Environ. Sci. Pollut. Res. Int. 2018, 25, 33391–33401. [Google Scholar] [CrossRef] [Green Version]
- Adefisoye, M.A.; Nwodo, U.U.; Green, E.; Okoh, A.I. Quantitative PCR Detection and Characterisation of Human Adenovirus, Rotavirus and Hepatitis A Virus in Discharged Effluents of Two Wastewater Treatment Facilities in the Eastern Cape, South Africa. Food Environ. Virol. 2016, 8, 262–274. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Hamilton, K.A.; Lobos, A.; Hughes, B.; Staley, C.; Sadowsky, M.J.; Harwood, V.J. Quantitative microbial risk assessment of microbial source tracking markers in recreational water contaminated with fresh untreated and secondary treated sewage. Environ. Int. 2018, 117, 243–249. [Google Scholar] [CrossRef]
- Ding, N.; Craik, S.A.; Pang, X.; Lee, B.; Neumann, N.F. Assessing UV Inactivation of Adenovirus 41 Using Integrated Cell Culture Real-Time qPCR/RT-qPCR. Water Environ. Res. 2017, 89, 323–329. [Google Scholar] [CrossRef]
- Mackowiak, M.; Leifels, M.; Hamza, I.A.; Jurzik, L.; Wingender, J. Distribution of Escherichia coli, coliphages and enteric viruses in water, epilithic biofilms and sediments of an urban river in Germany. Sci. Total Environ. 2018, 626, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Wyer, M.D.; Wyn-Jones, A.P.; Kay, D.; Au-Yeung, H.K.C.; Girones, R.; Lopez-Pila, J.; Husman, A.M.D.; Rutjes, S.; Schneider, O. Relationships between human adenoviruses and faecal indicator organisms in European recreational waters. Water Res. 2012, 46, 4130–4141. [Google Scholar] [CrossRef] [PubMed]
- Petterson, S.; Grondahl-Rosado, R.; Nilsen, V.; Myrmel, M.; Robertson, L.J. Variability in the recovery of a virus concentration procedure in water: Implications for QMRA. Water Res. 2015, 87, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lazaro, D.; Cook, N.; Ruggeri, F.M.; Sellwood, J.; Nasser, A.; Nascimento, M.S.; D’Agostino, M.; Santos, R.; Saiz, J.C.; Rzezutka, A.; et al. Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 2012, 36, 786–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhipi-Shrestha, G.; Hewage, K.; Sadiq, R. Microbial quality of reclaimed water for urban reuses: Probabilistic risk-based investigation and recommendations. Sci. Total Environ. 2017, 576, 738–751. [Google Scholar] [CrossRef]
- Hudu, S.A.; Alshrari, A.S.; Syahida, A.; Sekawi, Z. Cell Culture, Technology: Enhancing the Culture of Diagnosing Human Diseases. J. Clin. Diagn. Res. 2016, 10, De1–De5. [Google Scholar] [CrossRef]
- Shin, G.A.; Sobsey, M.D. Inactivation of norovirus by chlorine disinfection of water. Water Res. 2008, 42, 4562–4568. [Google Scholar] [CrossRef]
- Pecson, B.M.; Martin, L.V.; Kohn, T. Quantitative PCR for Determining the Infectivity of Bacteriophage MS2 upon Inactivation by Heat, UV-B Radiation, and Singlet Oxygen: Advantages and Limitations of an Enzymatic Treatment To Reduce False-Positive Results. Appl. Environ. Microbiol. 2009, 75, 5544–5554. [Google Scholar] [CrossRef] [Green Version]
- Hata, A.; Kitajima, M.; Katayama, H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J. Appl. Microbiol. 2013, 114, 545–554. [Google Scholar] [CrossRef]
- Leifels, M.; Hamza, I.A.; Krieger, M.; Wilhelm, M.; Mackowiak, M.; Jurzik, L. From Lab to Lake—Evaluation of Current Molecular Methods for the Detection of Infectious Enteric Viruses in Complex Water Matrices in an Urban Area. PLoS ONE 2016, 11, e0167105. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; Khezri, M.; Ollivier, J.; Le Guyader, F.S.; Rodriguez-Diaz, J.; Aznar, R.; Sanchez, G. Optimization of PMAxx pretreatment to distinguish between human norovirus with intact and altered capsids in shellfish and sewage samples. Int. J. Food Microbiol. 2018, 266, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nocker, A.; Cheung, C.Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B.; Adams, R.I.; Roman, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrodinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Seo, D.J.; Seo, J.; Oh, H.; Jeon, S.B.; Ha, S.D.; Myoung, J.; Choi, I.S.; Choi, C. Detection of viable murine norovirus using the plaque assay and propidium-monoazide-combined real-time reverse transcription-polymerase chain reaction. J. Virol. Methods 2015, 221, 57–61. [Google Scholar] [CrossRef]
- Karim, M.R.; Fout, G.S.; Johnson, C.H.; White, K.M.; Parshionikar, S.U. Propidium monoazide reverse transcriptase PCR and RT-qPCR for detecting infectious enterovirus and norovirus. J. Virol. Methods 2015, 219, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Ko, G. Using propidium monoazide to distinguish between viable and nonviable bacteria, MS2 and murine norovirus. Lett. Appl. Microbiol. 2012, 55, 182–188. [Google Scholar] [CrossRef]
- Bae, S.; Wuertz, S. Survival of host-associated bacteroidales cells and their relationship with Enterococcus spp., Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and adenovirus in freshwater microcosms as measured by propidium monoazide-quantitative PCR. Appl. Environ. Microbiol. 2012, 78, 922–932. [Google Scholar] [CrossRef]
- Quijada, N.M.; Fongaro, G.; Barardi, C.R.M.; Hernandez, M.; Rodriguez-Lazaro, D. Propidium Monoazide Integrated with qPCR Enables the Detection and Enumeration of Infectious Enteric RNA and DNA Viruses in Clam and Fermented Sausages. Front. Microbiol. 2016, 7, 2008. [Google Scholar] [CrossRef] [Green Version]
- Marie, V.; Lin, J. Viruses in the environment—Presence and diversity of bacteriophage and enteric virus populations in the Umhlangane River, Durban, South Africa. J. Water Health 2017, 15, 966–981. [Google Scholar] [CrossRef]
- Fraisse, A.; Niveau, F.; Hennechart-Collette, C.; Coudray-Meunier, C.; Martin-Latil, S.; Perelle, S. Discrimination of infectious and heat-treated norovirus by combining platinum compounds and real-time RT-PCR. Int. J. Food Microbiol. 2018, 269, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Werneck, L.M.C.; Vieira, C.B.; Fumian, T.M.; Caetano, T.B.; dos Santos, J.E.; Ferreira, F.C.; Pimenta, M.M.; Miagostovich, M.P. Dissemination of gastroenteric viruses in the production of lettuce in developing countries: A public health concern. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Delgado-Gardea, M.C.E.; Tamez-Guerra, P.; Gomez-Flores, R.; Mendieta-Mendoza, A.; de la Serna, F.J.Z.D.; Contreras-Cordero, J.F.; Erosa-de La Vega, G.; Perez-Recoder, M.C.; Sanchez-Ramirez, B.; Gonzalez-Horta, C.; et al. Prevalence of Rotavirus Genogroup A and Norovirus Genogroup II in Bassaseachic Falls National Park SurfaceWaters in Chihuahua, Mexico. Int. J. Environ. Res. Public Health 2017, 14, 482. [Google Scholar] [CrossRef] [PubMed]
- Prevost, B.; Goulet, M.; Lucas, F.S.; Joyeux, M.; Moulin, L.; Wurtzer, S. Viral persistence in surface and drinking water: Suitability of PCR pre-treatment with intercalating dyes. Water Res. 2016, 91, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Raurich, S.; Garriga, M.; Aymerich, T. Effect of Amplicon Length in Propidium Monoazide Quantitative PCR for the Enumeration of Viable Cells of Salmonella in Cooked Ham. Food Anal. Methods 2013, 6, 683–690. [Google Scholar] [CrossRef]
- Hamza, I.A.; Bibby, K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods 2019, 266, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Hernroth, B.E.; Conden-Hansson, A.C.; Rehnstam-Holm, A.S.; Girones, R.; Allard, A.K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: The first Scandinavian report. Appl. Environ. Microbiol. 2002, 68, 4523–4533. [Google Scholar] [CrossRef] [PubMed]
- Heim, A.; Ebnet, C.; Harste, G.; Pring-Akerblom, P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003, 70, 228–239. [Google Scholar] [CrossRef]
- Leifels, M.; Jurzik, L.; Wilhelm, M.; Hamza, I.A. Use of ethidium monoazide and propidium monoazide to determine viral infectivity upon inactivation by heat, UV- exposure and chlorine. Int. J. Hyg. Environ. Health 2015, 218, 686–693. [Google Scholar] [CrossRef]
- Strathmann, M.; Horstkott, M.; Koch, C.; Gayer, U.; Wingender, J. The River Ruhr—An urban river under particular interest for recreational use and as a raw water source for drinking water: The collaborative research project “Safe Ruhr”—Microbiological aspects. Int. J. Hyg. Environ. Health 2016, 219, 643–661. [Google Scholar] [CrossRef]
- Katayama, H.; Shimasaki, A.; Ohgaki, S. Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002, 68, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Hamza, I.A.; Jurzik, L.; Stang, A.; Sure, K.; Uberla, K.; Wilhelm, M. Detection of human viruses in rivers of a densly-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 2009, 43, 2657–2668. [Google Scholar] [CrossRef] [PubMed]
- Hennechart-Collette, C.; Martin-Latil, S.; Guillier, L.; Perelle, S. Determination of which virus to use as a process control when testing for the presence of hepatitis A virus and norovirus in food and water. Int. J. Food Microbiol. 2015, 202, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; Klemm, U.; Mas Marques, A.; Schreier, E. Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch. Virol. 2007, 152, 1709–1719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ye, C.; Lin, W.; Yu, X. Response to Comment on “UV Disinfection Induces a VBNC State in Escherichia coli and Pseudomonas aeruginosa”. Environ. Sci. Technol. 2015, 49, 10752–10753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ye, C.; Lin, H.; Lv, L.; Yu, X. UV disinfection induces a VBNC state in Escherichia coli and Pseudomonas aeruginosa. Environ. Sci. Technol. 2015, 49, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Sangsanont, J.; Katayama, H.; Kurisu, F.; Furumai, H. Capsid-Damaging Effects of UV Irradiation as Measured by Quantitative PCR Coupled with Ethidium Monoazide Treatment. Food Environ. Virol. 2014, 6, 269–275. [Google Scholar] [CrossRef]
- McLellan, N.L.; Lee, H.; Habash, M.B. Evaluation of propidium monoazide and long-amplicon qPCR as an infectivity assay for coliphage. J. Virol. Methods 2016, 238, 48–55. [Google Scholar] [CrossRef]
- Kim, S.-H.; Shahbaz, H.M.; Park, D.; Chun, S.; Lee, W.; Oh, J.-W.; Lee, D.-U.; Park, J. A combined treatment of UV-assisted TiO2 photocatalysis and high hydrostatic pressure to inactivate internalized murine norovirus. Innov. Food Sci. Emerg. Technol. 2017, 39, 188–196. [Google Scholar] [CrossRef]
- Girones, R.; Carratala, A.; Calgua, B.; Calvo, M.; Rodriguez-Manzano, J.; Emerson, S. Chlorine inactivation of hepatitis E virus and human adenovirus 2 in water. J. Water Health 2014, 12, 436–442. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-W.; Yoon, S.-R.; Lee, H.-M.; Lee, J.Y.; Kim, S.H.; Ha, J.-H. Use of RT-qPCR with combined intercalating dye and sodium lauroyl sarcosinate pretreatment to evaluate the virucidal activity of halophyte extracts against norovirus. Food Control 2019, 98, 100–106. [Google Scholar] [CrossRef]
- Lopez-Galvez, F.; Randazzo, W.; Vasquez, A.; Sanchez, G. Irrigating Lettuce with Wastewater Effluent: Does Disinfection with Chlorine Dioxide Inactivate Viruses? J. Environ. Qual. 2018, 47, 1139–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parshionikar, S.; Laseke, I.; Fout, G.S. Use of propidium monoazide in reverse transcriptase PCR to distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 2010, 76, 4318–4326. [Google Scholar] [CrossRef] [PubMed]
- Canh, V.D.; Kasuga, I.; Furumai, H.; Katayama, H. Viability RT-qPCR Combined with Sodium Deoxycholate Pre-treatment for Selective Quantification of Infectious Viruses in Drinking Water Samples. Food Environ. Virol. 2019, 11, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; Piqueras, J.; Rodriguez-Diaz, J.; Aznar, R.; Sanchez, G. Improving efficiency of viability-qPCR for selective detection of infectious HAV in food and water samples. J. Appl. Microbiol. 2018, 124, 958–964. [Google Scholar] [CrossRef]
- Gyawali, P.; Hewitt, J. Detection of Infectious Noroviruses from Wastewater and Seawater Using PEMAXTM Treatment Combined with RT-qPCR. Water 2018, 10, 841. [Google Scholar] [CrossRef]
- Monteiro, S.; Santos, R. Enzymatic and viability RT-qPCR assays for evaluation of enterovirus, hepatitis A virus and norovirus inactivation: Implications for public health risk assessment. J. Appl. Microbiol. 2018, 124, 965–976. [Google Scholar] [CrossRef]
- Petterson, S.R.; Ashbolt, N.J. QMRA and water safety management: Review of application in drinking water systems. J. Water Health 2016, 14, 571–589. [Google Scholar] [CrossRef]
- Kontchou, J.A.; Nocker, A. Optimization of viability qPCR for selective detection of membrane-intact Legionella pneumophila. J. Microbiol. Methods 2019, 156, 68–76. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Tellabati, M.; Nelli, R.K.; White, G.A.; Perez, B.B.; Sebastian, S.; Slomka, M.J.; Brookes, S.M.; Brown, I.H.; Dunham, S.P.; et al. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012, 9, 230. [Google Scholar] [CrossRef]
- Ryu, H.; Cashdollar, J.L.; Fout, G.S.; Schrantz, K.A.; Hayes, S. Applicability of integrated cell culture quantitative PCR (ICC-qPCR) for the detection of infectious adenovirus type 2 in UV disinfection studies. J. Environ. Sci. Health A 2015, 50, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Scaturro, M.; Fontana, S.; Dell’eva, I.; Helfer, F.; Marchio, M.; Stefanetti, M.V.; Cavallaro, M.; Miglietta, M.; Montagna, M.T.; De Giglio, O.; et al. A multicenter study of viable PCR using propidium monoazide to detect Legionella in water samples. Diagn. Microbiol. Infect. Dis. 2016, 85, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Katayama, H.; Kitajima, M.; Tohya, Y.; Ohgaki, S. Development of a real-time RT-PCR assay combined with ethidium monoazide treatment for RNA viruses and its application to detect viral RNA after heat exposure. Water Sci. Technol. 2011, 63, 502–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapron, C.D.; Ballester, N.A.; Fontaine, J.H.; Frades, C.N.; Margolin, A.B. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 2000, 66, 2520–2525. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Wang, S.W.; Jiang, S.C.; Bahnemann, J.; Hoffmann, M.R. Sunlight-Activated Propidium Monoazide Pretreatment for Differentiation of Viable and Dead Bacteria by Quantitative Real-Time Polymerase Chain Reaction. Environ. Sci. Technol. Lett. 2016, 3, 57–61. [Google Scholar] [CrossRef] [Green Version]
- Ruhrverband. Abschlussbericht zum Forschungsvorhaben Ertüchtigung Kommunaler Kläranlagen durch den Einsatz von Verfahren Mit UV-Behandlung (German); Ruhrverband: Essen, Germany, 2012. [Google Scholar]
- Seinige, D.; Krischek, C.; Klein, G.; Kehrenberg, C. Comparative analysis and limitations of ethidium monoazide and propidium monoazide treatments for the differentiation of viable and nonviable campylobacter cells. Appl. Environ. Microbiol. 2014, 80, 2186–2192. [Google Scholar] [CrossRef]
- Tondera, K.; Klaer, K.; Gebhardt, J.; Wingender, J.; Koch, C.; Horstkott, M.; Strathmann, M.; Jurzik, L.; Hamza, I.A.; Pinnekamp, J. Reducing pathogens in combined sewer overflows using ozonation or UV irradiation. Int. J. Hyg. Environ. Health 2015, 218, 731–741. [Google Scholar] [CrossRef]
- Tondera, K.; Klaer, K.; Koch, C.; Hamza, I.A.; Pinnekamp, J. Reducing pathogens in combined sewer overflows using performic acid. Int. J. Hyg. Environ. Health 2016, 219 Pt B, 700–708. [Google Scholar] [CrossRef]
- Kitajima, M.; Rachmadi, A.T.; Iker, B.C.; Haramoto, E.; Gerba, C.P. Genetically distinct genogroup IV norovirus strains identified in wastewater. Arch. Virol. 2016, 161, 3521–3525. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Qiu, Y.; Pang, X.L.; Ashbolt, N.J. Level of spiked virus necessary to correctly assess enteric virus recovery in water matrices. Appl. Environ. Microbiol. 2019. [Google Scholar] [CrossRef]
- Schoen, M.E.; Ashbolt, N.J.; Jahne, M.A.; Garland, J. Risk-based enteric pathogen reduction targets for non-potable and direct potable use of roof runoff, stormwater, and greywater. Microb. Risk Anal. 2017, 5, 32–43. [Google Scholar] [CrossRef]
- Oka, T.; Stoltzfus, G.T.; Zhu, C.; Jung, K.; Wang, Q.; Saif, L.J. Attempts to grow human noroviruses, a sapovirus, and a bovine norovirus in vitro. PLoS ONE 2018, 13, e0178157. [Google Scholar] [CrossRef] [PubMed]
- Van Abel, N.; Schoen, M.E.; Kissel, J.C.; Meschke, J.S. Comparison of Risk Predicted by Multiple Norovirus Dose-Response Models and Implications for Quantitative Microbial Risk Assessment. Risk Anal. 2017, 37, 245–264. [Google Scholar] [CrossRef] [PubMed]
- Langlet, J.; Kaas, L.; Croucher, D.; Hewitt, J. Effect of the Shellfish Proteinase K Digestion Method on Norovirus Capsid Integrity. Food Environ. Virol. 2018, 10, 151–158. [Google Scholar] [CrossRef] [PubMed]




| Protocol | Primer | Primers & Probe Sequences 5’–3’ | Amplicon (bp) | Gene Target | Ref. |
|---|---|---|---|---|---|
| HAdV-Heim | AQ1 | GCC ACG GTG GGG TTT CTA AAC TT | 139 | Hexon | [50] |
| AQ2 | GCC CCA GTG GTC TTA CAT GCA CAT C | ||||
| AdV-P | [Hex]-TGC ACC AGA CCC GGG CTC AGG TAC TCC GA-[BHQ1] | ||||
| HAdV-Hernroth | Ad.hex.up | CWT ACA TGC ACA TCK CSG G | 69 | [49] | |
| Ad.hex.do | CRC GGG CRA AYT GCA CCA G | ||||
| AdV-ACDEF | [6FAM]-CCG GGC TCA GGT ACT CCG AGG CGT CCT-[TAMRA] | ||||
| AdV-B | [6FAM]-CCG GAC TCA GGT ACT CCG AAG CAT CCT-[TAMRA] | ||||
| MNV | TMP 1 | AGA GGA ATC TAT GCG CCT GG | 92 | ORF2 | [56] |
| TMP 2 | GAA GGC GGC CAG AGA CCA C | ||||
| TMP | [6FAM]-GCC ACT CCG CAC AAA CAG CCC-[BHQ1] |
| False +/− for Detection of Infectious HAdV | |||
|---|---|---|---|
| Source | Assay | False + | False – |
| Surface (n = 51) | PMA ci-qPCR | 3 (5.89) | 5 (9.80) |
| EMA ci-qPCR | 4 (7.84) | 17 (33.33) | |
| qPCR | 39 (76.47) | 0 (0.00) | |
| Before UV (n = 12) | PMA ci-qPCR | 5 (41.67) | 1 (16.67) |
| EMA ci-qPCR | 4 (33.33) | 1 (16.67) | |
| qPCR | 8 (66.67) | 0 (0.00) | |
| After UV (n = 12) | PMA ci-qPCR | 2 (16.67) | 1 (8.33) |
| EMA ci-qPCR | 2 (16.67) | 2 (16.67) | |
| qPCR | 8 (66.67) | 3 (20.00) | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leifels, M.; Shoults, D.; Wiedemeyer, A.; Ashbolt, N.J.; Sozzi, E.; Hagemeier, A.; Jurzik, L. Capsid Integrity qPCR—An Azo-Dye Based and Culture-Independent Approach to Estimate Adenovirus Infectivity after Disinfection and in the Aquatic Environment. Water 2019, 11, 1196. https://doi.org/10.3390/w11061196
Leifels M, Shoults D, Wiedemeyer A, Ashbolt NJ, Sozzi E, Hagemeier A, Jurzik L. Capsid Integrity qPCR—An Azo-Dye Based and Culture-Independent Approach to Estimate Adenovirus Infectivity after Disinfection and in the Aquatic Environment. Water. 2019; 11(6):1196. https://doi.org/10.3390/w11061196
Chicago/Turabian StyleLeifels, Mats, David Shoults, Alyssa Wiedemeyer, Nicholas J. Ashbolt, Emanuele Sozzi, Angela Hagemeier, and Lars Jurzik. 2019. "Capsid Integrity qPCR—An Azo-Dye Based and Culture-Independent Approach to Estimate Adenovirus Infectivity after Disinfection and in the Aquatic Environment" Water 11, no. 6: 1196. https://doi.org/10.3390/w11061196